DNA helps reveal bat diets
What do carnivorous animals eat? Predation drives evolution and underlies ecology, yet except for a few easily observed species, it is surprisingly hard to determine what eats what. In June 2009 Mol Ecol, researchers from University of Guelph and University of Western Ontario, Canada, apply DNA testing to help solve diet of Eastern red bat Laiurus borealis. L. borealis is the commonest tree-roosting bat in North America, ranging from Canada and United States east of the Rocky Mountains into Central and northern South America. Like other insectivorous bats, L. borealis uses echolocation to detect night-flying insects. Many moth species have evolved “ears” that detect the ultrasonic sounds emitted by bats and exhibit defensive behaviors in response to echolocation signals, making bats and moths an interesting study in predator-prey co-evolution.
Clare and co-workers applied standardized DNA testing to insect parts in faecal samples collected from 56 mist-net trapped bats. Guano samples were frozen for up to 2 y then soaked in 95% ethanol for 12 h and examined with a dissecting microscope. Prey items including “legs, wings, antennae, eye cases, exoskeletal fragments, eggs” were isolated and stored separately in 96 well-plates. DNA extraction, amplification, and sequencing were performed using standard techniques and broad-range insect primers (LepF1/LepR1). COI sequences were compared to the 127,000 reference sequences of North American arthropods in BOLD database www.barcodinglife.org at the time of the study. Test sequences with >/=99% identity to reference sequence(s) and without equivalent similarity to other species in the database were given species-level identifications; those with less than 99% identity to reference sequence(s) were assigned to higher-level taxonomic categories.
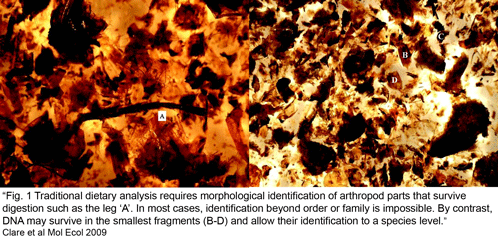 Clare et al obtained sequence data from 89% of 896 arthropod fragments; 78% of these were identified to species or genus level (the remaining 22% showed sequence similarity to bacteria, fungi, or were unidentifiable or chimeric), with a total of 127 prey species identified (125 insects, mainly lepidoptera including a number of economically important pest species, and 2 spiders). The “molecular scatology” approach documented greater diversity in prey species than prior studies based on morphologic analysis. Most prey were identified only once, with an average of 3.5 species per guano sample. Surprisingly, “more than 60% [of recovered insects] appear to have ears capable of hearing the echolocation hunting calls of L. borealis.” The authors speculate the abundance of eared moths might reflect bats hunting around streetlights, as moths in such brightly-lit environments are thought to use daytime predator-avoidance strategies rather than nocturnal responses to echolocation. There was a notable absence of actiid and tortricid moths, given their local abundance, suggesting these moths may have alternative predator-avoidance strategies.
Clare et al obtained sequence data from 89% of 896 arthropod fragments; 78% of these were identified to species or genus level (the remaining 22% showed sequence similarity to bacteria, fungi, or were unidentifiable or chimeric), with a total of 127 prey species identified (125 insects, mainly lepidoptera including a number of economically important pest species, and 2 spiders). The “molecular scatology” approach documented greater diversity in prey species than prior studies based on morphologic analysis. Most prey were identified only once, with an average of 3.5 species per guano sample. Surprisingly, “more than 60% [of recovered insects] appear to have ears capable of hearing the echolocation hunting calls of L. borealis.” The authors speculate the abundance of eared moths might reflect bats hunting around streetlights, as moths in such brightly-lit environments are thought to use daytime predator-avoidance strategies rather than nocturnal responses to echolocation. There was a notable absence of actiid and tortricid moths, given their local abundance, suggesting these moths may have alternative predator-avoidance strategies.
This study documents the diversity of L. borealis prey, and hints at how much more we will learn from broad application of standardized DNA analysis to food chains, including such unexpected findings as possible disruptive effects of man-made lighting on local ecosystems.
This entry was posted on Tuesday, June 9th, 2009 at 7:46 am and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.