Neotropical explorers map avian diversity
There are more bird species in the Neotropics than in any other biogeographic region (including many more as yet undescribed; see for example application of COI barcoding to cryptic diversity in Brazilian tyrant flycatchers Chaves et al Nov 2008 Mol Ecol Resources). Mapping this diversity with COI is both exciting and perhaps challenging. Some have wondered if the plethora of neotropical avian species might overwhelm the ability of a single mtDNA gene to resolve differences among species.
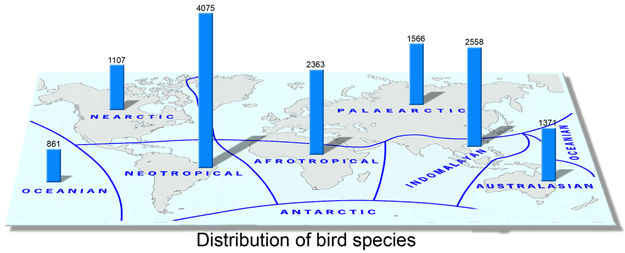
There is apparently no need to worry. In February 2009 PLoS ONE researchers from University of Guelph, Canada, and Museo Argentino de Ciencias Naturales, Argentina report on 1,594 COI barcode sequences from 500 species, which represents 51% of of Argentine birds. Kerr and colleagues find COI barcodes work here as elsewhere, ie most species show deep divergences from their “nearest neighbor” in the data set. Overall, 491 of 500 (98%) of species had distinct barcode(s). Of the nine species not resolved, six were Sporophila spp (common name “seedeaters”); these comprised a single cluster of shared barcodes. Although morphologically distinct, I wonder if these might be analogous to North America juncos, which have regional plumage variants without genetic differentiation. As recently as 1973 they were considered to be a complex of 5 species; these are now lumped into one, Junco hyemalis.
21 Argentine birds showed 2 or more distinct clusters (maximum intraspecific distance 1.52-5.41%). Do these represent different species? The authors genuflect to taxonomic tradition, ie “levels of genetic differentiation do not dictate taxonomic status” and gently suggest “barcode analysis illuminates those taxa and those segments of their ranges where further research is justified.”
I am impressed with the ease of analyzing combined data sets; in this case the scientists merged their Neotropical COI data with that of Nearctic birds (2,615 sequences/659 species). With a click of a button so to speak, Kerr and colleagues were able to determine that 10 of 42 (24%) species shared between the 2 regions showed large genetic differences, which common sense tells us indicate long-standing reproductive isolation (ie species status). Most of these North-South divergences were in plain-colored passerines or nocturnal species, and often with disjunct ranges consisting of a northern migratory and southern resident population. The former suggests why these might have been overlooked and the latter suggests how they arose. This work highlights how merging data sets amplifies the power of regional studies–one of the benefits of standardizing on a defined barcode mtDNA region, one that will grow with increasing size of barcode library.
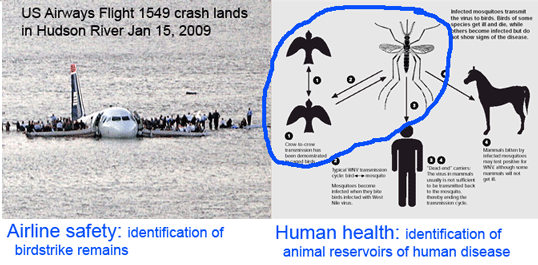
In addition to enabling rapid and low-cost mapping of avian diversity including discovery of divergent lineages, which in most cases are indicators of new species, avian DNA barcoding establishes a genetic reference library with practical benefits to society, as in examples illustrated above. The DNA barcoding effort including All Birds Barcoding Initiative (ABBI) draws interest from many persons who live outside of museum walls.
This entry was posted on Monday, February 16th, 2009 at 3:56 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.