“Tag sequencing” reveals vast microbial diversity
A “tag sequencing” approach analogous to mtCOI sequencing for barcoding multicellular organisms reveals vast numbers of very rare, highly divergent, deep sea microbes. In August 2006 PNAS (Sogin et al Proc Natl Acad Sci USA 32:12125), researchers from Marine Biological Laboratory at Woods Hole and Royal Netherlands Institute for Sea Research report on pooled bacterial samples collected at 550-1,710 meters in the Atlantic Ocean. To enable detection of rare populations, they focused on a short hypervariable region of 16s rRNA (only 79 bases) and analyzed a large number of PCR amplicons (118,000!) using 454 Life Sciences technology. This approach makes it economical to analyze enormous numbers of sequences from pooled environmental samples and avoids possible selection artifacts due to biases in amplifying longer PCR products and in cloning. Remarkably, the very short 79 base pair “tag” captured about 90% of the sequence differences in full-length 1500 base pair 16s rRNA sequences.
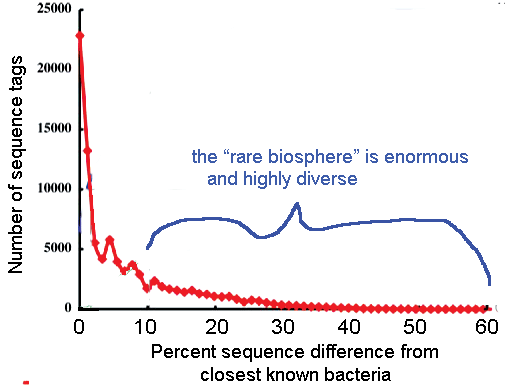
The results were compared to a V6 hypervariable region database, which contains about 40,000 unique V6 sequences extracted from the nearly 120,000 published bacterial rRNA gene sequences. A small number of sequence tags similar to known bacteria made up most of the samples, including 25% that were identical to sequences in the database and 40% that were no more than 3% different. Overall 75% of “total tags” were less than 10% different from previously sequenced bacteria. The remaining 25% was comprised of thousands of low abundance, extraordinarily diverse populations. The authors conclude the “rare biosphere” is “an ancient and..nearly inexhautible source of genomic innovation..[that] at different times in earth’s history..may have had a profound impact on shaping planetary processes.” There is a lot more we will learn through standardized genetic analysis using short sequences, including mtCOI barcodes and v6 rRNA tags, applied to vast numbers of organisms.
This entry was posted on Saturday, August 19th, 2006 at 10:54 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.
September 27th, 2006 at 2:05 pm
[…] Since there are about 1.1 million named multicellular animal species, “species identification” is a vast area for scientific research and practical application of DNA barcoding. It seems likely that the 1.1 million known species includes most of the more abundant and wide-ranging species, and most that are of direct economic or scientific importance to humans. It is generally believed there are many more undescribed species than what has already been named. These may be largely rarer species with limited distributions (see earlier post on “rare microbial biosphere“). Population sizes and ranges in the undescribed biosphere, together with measures of genetic diversity (see last week’s post) might be interesting research areas. […]