Counting zooplankton diversity with DNA
 Marine zooplankton comprise an enormous mass of diverse organisms distributed throughout the world’s oceans from deep waters to surface. Zooplankton include representatives of at least dozen phyla, some of which are larval forms of much larger animals, and challenge identification with their diversity and tiny size. In current BMC Genomics (open access) researchers from University of Tokyo and Osaka Medical College, as part of Census of Marine Zooplankton (CMarZ) program of the Census of Marine Life (CoML), apply single-gene sequencing to the task. Machida and colleagues collected at a Micronesia site using a single pass with 2m^2 plankton net from depth of 721 meters to surface, obtaining 60 mL of of zooplankton (large organisms, up to 4 cm, were discarded). Rather than direct DNA sequencing, the researchers isolated mRNA from the pooled sample and constructed a cDNA library from which they analyzed 1,336 inserts. The rationale for these extra steps was to avoid sequencing pseudogenes present in genomic DNA (but not transcribed into mRNA). It would be interesting to know if this strategy was based on experience or is a theoretical precaution.
Marine zooplankton comprise an enormous mass of diverse organisms distributed throughout the world’s oceans from deep waters to surface. Zooplankton include representatives of at least dozen phyla, some of which are larval forms of much larger animals, and challenge identification with their diversity and tiny size. In current BMC Genomics (open access) researchers from University of Tokyo and Osaka Medical College, as part of Census of Marine Zooplankton (CMarZ) program of the Census of Marine Life (CoML), apply single-gene sequencing to the task. Machida and colleagues collected at a Micronesia site using a single pass with 2m^2 plankton net from depth of 721 meters to surface, obtaining 60 mL of of zooplankton (large organisms, up to 4 cm, were discarded). Rather than direct DNA sequencing, the researchers isolated mRNA from the pooled sample and constructed a cDNA library from which they analyzed 1,336 inserts. The rationale for these extra steps was to avoid sequencing pseudogenes present in genomic DNA (but not transcribed into mRNA). It would be interesting to know if this strategy was based on experience or is a theoretical precaution.
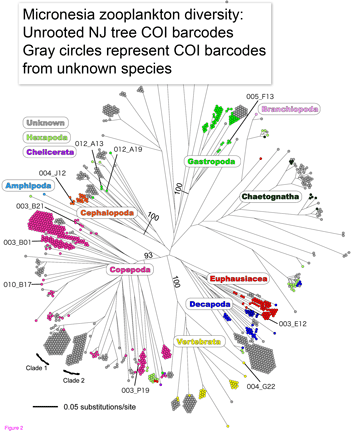 Machida and colleagues found evidence for 189 species, only 10 of which could be confidently matched to reference sequences. This report demonstrates that this sort of “kitchen blender” approach, which has previously been applied largely to bacterial and archaeal communities, shows promise for assemblages of eukaryotes and reveals surprisingly few organisms have reference sequences in databases. Identified organisms included several copepods as well as presumably larval forms of Sthenoteuthis oualaniensis (Purple-back flying squid) and Coryphaena hippurus (Common dolphinfish)!
Machida and colleagues found evidence for 189 species, only 10 of which could be confidently matched to reference sequences. This report demonstrates that this sort of “kitchen blender” approach, which has previously been applied largely to bacterial and archaeal communities, shows promise for assemblages of eukaryotes and reveals surprisingly few organisms have reference sequences in databases. Identified organisms included several copepods as well as presumably larval forms of Sthenoteuthis oualaniensis (Purple-back flying squid) and Coryphaena hippurus (Common dolphinfish)!
Species identification by DNA opens major avenues for for ecosystem research. The NJ tree at left suggests that even in absence of close matches, 500 bp of mtDNA is sufficient to sort most specimens into appropriate higher-level groups. To better understand the changing oceans, we need biological monitoring machines akin to physical instruments for studying weather and climate, which routinely monitor thousands of sites. It seems to me the only practical way to monitor biological “weather” is by repeatedly sampling species assemblages at multiple points, and particularly in aqueous environments, automated species identification with DNA will be an important analytic method.
This entry was posted on Sunday, September 20th, 2009 at 6:33 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.