DNA helps sort out really big animals, crowding Ark
 How many giraffes were onboard the Ark? Giraffes are classified as a single species, Giraffa camelopardalis, with five to nine subspecies proposed based on regional variation in pelage (coat pattern). In 21 dec 2007 BMC Biology (open access) researchers from University of California, Los Angeles; Center for Conservation Research, Omaha Zoo; and Mpala Research Centre, Kenya, investigate genetic variation in giraffes across African continent.
How many giraffes were onboard the Ark? Giraffes are classified as a single species, Giraffa camelopardalis, with five to nine subspecies proposed based on regional variation in pelage (coat pattern). In 21 dec 2007 BMC Biology (open access) researchers from University of California, Los Angeles; Center for Conservation Research, Omaha Zoo; and Mpala Research Centre, Kenya, investigate genetic variation in giraffes across African continent.
Using biopsy darts, the authors collected skin specimens from 266 giraffes at 19 localities in West, East, and South Africa. A 654 nucleotide region of mtDNA spanning cytb and control sequences was analyzed, revealing 35 haplotypes, and the remainder of the cytb gene (1709 bp total) was sequenced from one individual from each of the 35 haplotypes. The mtDNA sequences clustered into six reciprocally monophyletic lineages, which corresponded to groupings according to pelage pattern and regional location, and were largely concordant with subspecies designations. Genetic distances suggested these groups have been reproductively isolated for 0.3 to 1.6 MY, similar to calculated divergence times among other closely-related mammals.
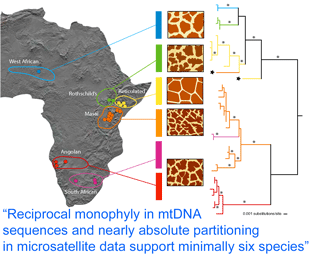 Analysis of 14 nuclear microsatellites from 381 individuals at 18 locations (it is not clear whether these are the same individuals as above) recovered the same six groups and suggested additional genetic subdivisions within some groups. Although at least some of the genetically and pelage-defined clusters have overlapping or adjacent ranges without geographic barriers, only three (0.8%) of individuals were identified as hybrids. These findings raise interesting questions about giraffe biology; for example, is there behavioral isolation perhaps based on visual recognition of pelage patterns?
Analysis of 14 nuclear microsatellites from 381 individuals at 18 locations (it is not clear whether these are the same individuals as above) recovered the same six groups and suggested additional genetic subdivisions within some groups. Although at least some of the genetically and pelage-defined clusters have overlapping or adjacent ranges without geographic barriers, only three (0.8%) of individuals were identified as hybrids. These findings raise interesting questions about giraffe biology; for example, is there behavioral isolation perhaps based on visual recognition of pelage patterns?
It is impressive that species can be overlooked in such large, boldly patterned, iconic animals. Might there be similar divisions within the numerous species of small, brown, rarely seen mammals? Routine DNA analysis of a standardized mtDNA region (aka DNA barcoding) will help discover how finely divided animal biodiversity is. Wilson and Reeder’s Mammal Species of the World, Third Edition lists 5,419 species, so this appears to be an achievable goal for our mammalian kin (list available online https://nmnhgoph.si.edu/msw/). I hope the authors include barcode region COI in their next analyses, so their data can be easily combined with other data sets, including the 28,560 mammalian barcode records in BOLD to date.
This entry was posted on Friday, June 13th, 2008 at 7:42 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.