DNA barcodes suggest fractal nature of genome
 Growing data sets demonstrate DNA barcoding usually works, but why? Why does a very short stretch of DNA, such as a DNA barcode which usually represents less than one one-millionth of the genome, enable identification of most animal species? In computer language, Rod Page describes a DNA barcode as “embedded metadata“. Here I suggest an analogy to fractals, which might help convey what DNA barcodes reveal about how genomes are constructed.
Growing data sets demonstrate DNA barcoding usually works, but why? Why does a very short stretch of DNA, such as a DNA barcode which usually represents less than one one-millionth of the genome, enable identification of most animal species? In computer language, Rod Page describes a DNA barcode as “embedded metadata“. Here I suggest an analogy to fractals, which might help convey what DNA barcodes reveal about how genomes are constructed.
DNA barcoding usually works because patterns seen in very short DNA sequences usually reflect patterns seen in longer sequences. In this way, DNA barcodes demonstrate “self-similarity”, a fundamental property of fractals. In March 28, 2007 PLoS One, researchers from Concordia University, Quebec, analyze 849 complete animal mitochondrial genomes, comparing GC composition in 648 bp COI barcode region to GC composition in the mitochondrial genome as a whole. Min and Hickey found “such short sequences can yield important, and surprisingly accurate, information about the [mitochondrial] genome as a whole. In other words, for unsequenced genomes, 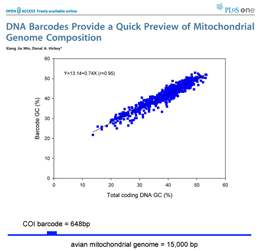 the DNA barcodes can provide a quick preview of the whole genome.” It will be of great interest to extend this analysis to compare mitochondrial barcodes to nuclear genomes; the general success of barcoding approach suggests there will be similarly close correlation.
the DNA barcodes can provide a quick preview of the whole genome.” It will be of great interest to extend this analysis to compare mitochondrial barcodes to nuclear genomes; the general success of barcoding approach suggests there will be similarly close correlation.
Overall, the patterning of barcode differences supports the emerging view that selective sweeps prune mitochondrial diversity within species and mitochondrial and nuclear co-evolution are tightly linked.
.
.
.
.
This entry was posted on Saturday, May 26th, 2007 at 10:07 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.
May 29th, 2007 at 9:55 pm
Interesting – I was not aware that small sections of DNA exhibit self-similarity. Yet another area to apply quantum methods to…
June 6th, 2007 at 5:06 pm
I’m not sure this applies. Note that the fractal nature of a part of a fractal image can de determined by examination of both the smaller details and the larger details. This is not the case with barcodes, where the smaller details of the sequence do not reflect exactly the organization of the whole genome. It is like saying that tossing a coin a few times is representative of many tosses. One cannot predict the long frequency data from a particular coin from only a few tosses. One needs sufficient data to do a chi-square test. Same with barcode data.
June 25th, 2007 at 2:53 pm
Here comes FractoGene (http://www.fractogene.com) – with the collapse of both “Genes” thesis and “JunkDNA” antithesis (with “genes” appearing obviously fragmented and the “Junk” proving to be anything, but), fractal properties (such as “barcode”) are found both in the DNA and are visible e.g. in the arborization of the fern.
Another isntance of “self-similarity” is represented by the onion – though some species of onions have enormously greater amount of formerly “Junk” DNA than others.
Very loose and mostly peri-scientific blogs, such as Panda’s thumb http://www.pandasthumb.org/archives/2007/06/junk_dna_junk_s.html are on record now to measure our understanding of the function of the whole genome by the ability to “pass the onion test”, best formulated by Ryan Gregory (essentially the same as Richard Dawkins’ “Salamander paradox”; that different sub-species of Salamandra have a great diversity in their size of “junk DNA”).
Thus far, FractoGene came forward with an explanation – awaiting other theoretical accounts …
pellionisz_at_junkdna.com