Voucher and collection information in GenBank records
A core tenet of DNA barcoding initiative, beginning with the first workshops in 2003, is that reference sequences should be linked to vouchered specimens stored in museums, so that data can be re-checked. This also provides visibility to collections. For example, “GenBank DQ433554 Crotophaga ani voucher KU 89123 cytochrome oxidase subunit 1 (COI) gene, partial cds; mitochondrial” contains voucher information in the title and the record itself, at least for those who know “KU” refers to University of Kansas. The GenBank file contains a “LinkOut” to the BOLD page which spells out the collection name. The GenBank file (and the BOLD record) could also include a “LinkOut” to the museum itself, although I do not find examples of this feature being used.
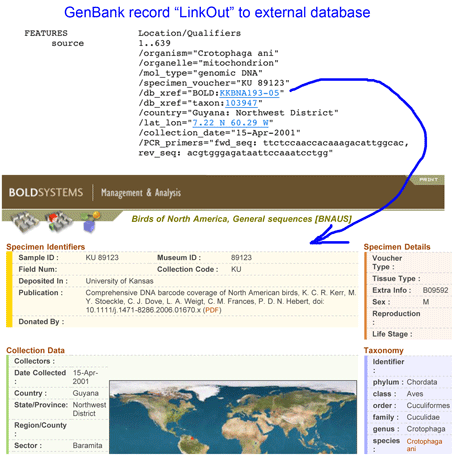
More generally, is collection information available in GenBank records? Taking birds as an example, there are 475,273 GenBank avian records; eliminating the five most-represented species (Chicken, Turkey, Mallard, Zebra Finch, Fairy Wren) leaves 108,766 sequences, of which about half (48,915) contain the word “voucher.” This sounds promising but my unscientific sample suggests most entries in the “voucher” field are cryptic designations that do not identify the institution storing the specimen. I tried searching by acronyms for some of the larger collections. Louisiana State University has the largest avian tissue collection in the world with about 40,000 specimens; searching “LSU AND aves[organism] AND voucher” returned only 1,148 records, which seems likely to underrepresent the museum’s contribution. Results for some other large collections were higher but still appear to be incorrectly small considering there are 100,000+ avian GenBank records: (Burke Museum (UWBM) 3,318; Field Museum (FMNH), 2,593; American Museum of Natural History (AMNH), 1,994; Smithsonian (USNM), 1,920; University of Kansas (KU), 684 records).
I conclude that researchers and collections will benefit from following practices promoted by DNA barcode initiative for GenBank records including taking advantage of GenBank’s “LinkOut” feature.
This entry was posted on Friday, July 17th, 2009 at 12:28 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.
July 20th, 2009 at 9:51 pm
This is an interesting post and your efforts to sleuth collection records from GenBank entries are commendable. It is noteworthy that the barcoding community successfully lobbied for the addition of new fields to the INSDC (DDBJ, EMBL & GenBank) feature table to accommodate a structured reference to voucher specimens, GPS coordinates, name of collector, name of specimen identifier, etc. and these fields are now available for any sequence submission. Sadly, with the notable exception of Arctos, few museum databases yet support external linkage with GenBank or BOLD. Even fewer of them support dynamic web services… for those that even have online collection catalogs. Sadly it will be a long time before collection records are digitized and brought into the e-Biosphere, particularly for invertebrates. Even when such collection records are available online thru portals such as GBIF, without barcodes to qualify the application of Linnean names associated with each record it is difficult to interpret the validity of the results obtained…
July 25th, 2009 at 7:44 pm
This is helpful reminder that “voucher” and related fields now available in GenBank are outcome of barcode initiative, with general benefit to collection and biodiversity science communities, and that there is a lot-of-opportunity/long-way-to-go to unlock scientific and social value of collections.