Solving puzzles of mitochondrial variation within and among species
What limits mitochondrial variation within species? In January 2008 PLoS Biology researchers from Karolinksa Institute, Sweden, and University of Newcastle upon Tyne, United Kingdom, report on an ingenious mouse model that shows strong purifying selection acting within a single generation, or even earlier, during embryogenesis. Stewart and colleagues employed “mtDNA mutator” mice which are homozygous defective for a nuclear gene which encodes a proof-reading subunit of mtDNA polymerase. These mice have increased levels of mtDNA mutations in all tissues, with mutations evenly distributed along all codon positions in mtDNA protein genes, accelerated senescence and “a number of phenotypes associated with mitochondrial diseases.” mtDNA mutator mice were backcrossed to wild-type mice to produce offspring that inherited defective mitochondria but whose nuclear genome is homozygous normal at the mtDNA polymerase locus. 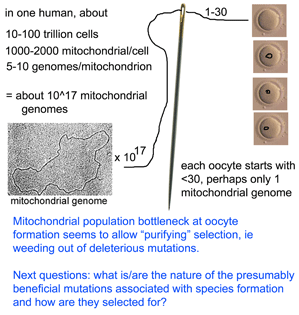 They then sequenced entire mitochondrial genomes from 190 of these progeny individuals in N2 to N6 generations (N2 is the first backcross that is homozygous normal at mtDNA mutator locus). To skip to the conclusion, most of the non-synonomous mitochondrial mutations were eliminated, leaving a pattern of excess synonymous mutations similar to that seen in human populations (which are the largest dataset so far for mitochondrial variation). The authors conclude that the mitochondrial population bottleneck known to occur at oogenesis, which deposits just one or few mitochondrial genomes per oocyte, means each mitochondrial genome must stand on its own so to speak, with the result that those eggs, embryos, or offspring harboring defective mitochondria will fail to survive. My figure at right tries to illustrate part of this process.
They then sequenced entire mitochondrial genomes from 190 of these progeny individuals in N2 to N6 generations (N2 is the first backcross that is homozygous normal at mtDNA mutator locus). To skip to the conclusion, most of the non-synonomous mitochondrial mutations were eliminated, leaving a pattern of excess synonymous mutations similar to that seen in human populations (which are the largest dataset so far for mitochondrial variation). The authors conclude that the mitochondrial population bottleneck known to occur at oogenesis, which deposits just one or few mitochondrial genomes per oocyte, means each mitochondrial genome must stand on its own so to speak, with the result that those eggs, embryos, or offspring harboring defective mitochondria will fail to survive. My figure at right tries to illustrate part of this process.
In the same issue, David Rand, Brown University, provides a lucid commentary on Stewart et al’s research putting it in the context of mitochondrial and evolutionary biology, and suggesting next steps. Among others, he notes “the new mouse study also begs new questions about positive selection on mtDNA. …it is interesting that no signature of a selective sweep leading to fixation of a novel mtDNA variant was evident in the data”.
Purifying selection against deleterious mutations enabled by an embryonic bottleneck may save mtDNA from “mutational meltdown”. Now we need to understand more about the positive selection on mtDNA that presumably occurs when species adapt to new environments or diverge. I believe that growing mtDNA databases in the form of COI barcodes from a diversity of organisms with varying size, lifespan, population size, and reproductive strategy, in a diversity of environments including marine, terrestrial, temperature, and tropical regions will help solve this puzzle.
This entry was posted on Wednesday, February 27th, 2008 at 12:32 am and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.
February 27th, 2008 at 12:22 pm
This model suggests there might be selection at the level of the bottleneck itself. According to this idea, mitochondria and host are in competition, with many mitochondria trying to get into the egg, and the host acting as a gatekeeper, trying to let in just one or a few. Like a host-parasite interaction, this could be a continuous arms race that leads to rapid co-evolutionary change.
February 27th, 2008 at 3:11 pm
Thank-you for the interest in our work. The area has been producing some interesting results of late. Here are some other recent articles.
Another germline selection experiment.
Eric A. Shoubridge and Timothy Wai PERSPECTIVE: MEDICINE: Sidestepping Mutational Meltdown Science 2008 319, (5865) 914-915.
Weiwei Fan, Katrina G. Waymire, Navneet Narula, Peng Li, Christophe Rocher, Pinar E. Coskun, Mani A. Vannan, Jagat Narula, Grant R. MacGregor, and Douglas C. Wallace
A Mouse Model of Mitochondrial Disease Reveals Germline Selection Against Severe mtDNA Mutations Science 2008 319, (5865) 958-962.
Redefining the bottleneck…
Cree LM, Samuels DC, de Sousa Lopes SC, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HH, Chinnery PF. “A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes.” Nat Genet. 2008 40(2):249-54.
Cao L, Shitara H, Horii T, Nagao Y, Imai H, Abe K, Hara T, Hayashi J, Yonekawa H.
“The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells.” Nat Genet. 2007 39(3):386-90.