Learning about lichens with DNA
June 1st, 2011
 In 1867 Swiss botanist Simon Schwendener was the first to recognize that lichens were symbiotic associations of fungi and algae (or, as subsequently discovered, cyanobacteria) (for more info, try this EOL podcast on lichen–“a tropical rainforest in miniature”). Today about 13,500 species are described (lichens are named for fungal component), representing 18% of the 74,000 known fungi. It is remarkable that so few fungi have been named, given that estimated diversity is 1.5 million (Hawksworth, 2001). This presumably reflects difficulty of morphologic diagnosis of often microscopic, unculturable organisms with diverse life forms and highlights a need for molecular methods. Several recent epidemics causing serious animal and plant mortality have turned out to be newly recongized fungi [including Batrochochytrium dendrobatidis (chytridiomycosis in amphibians), Geomyces destructans (White-nose Syndrome in bats), Cryophonectria parasitica (Chestnut blight), and Ophiostoma spp. (Dutch elm disease)], hinting at the hidden diversity and importance of fungi.
In 1867 Swiss botanist Simon Schwendener was the first to recognize that lichens were symbiotic associations of fungi and algae (or, as subsequently discovered, cyanobacteria) (for more info, try this EOL podcast on lichen–“a tropical rainforest in miniature”). Today about 13,500 species are described (lichens are named for fungal component), representing 18% of the 74,000 known fungi. It is remarkable that so few fungi have been named, given that estimated diversity is 1.5 million (Hawksworth, 2001). This presumably reflects difficulty of morphologic diagnosis of often microscopic, unculturable organisms with diverse life forms and highlights a need for molecular methods. Several recent epidemics causing serious animal and plant mortality have turned out to be newly recongized fungi [including Batrochochytrium dendrobatidis (chytridiomycosis in amphibians), Geomyces destructans (White-nose Syndrome in bats), Cryophonectria parasitica (Chestnut blight), and Ophiostoma spp. (Dutch elm disease)], hinting at the hidden diversity and importance of fungi.
Back to lichens–in March 2011 New Phytologist, researchers from Royal Botanic Garden (RBG) at Edinburgh and Kew report on DNA barcoding of lichenized fungi using internal transcribed spacer (ITS) region. ITS has been widely used in fungal taxonomy and has been proposed as a standard barcode region for this group (the standard barcode for animals, COI, has so far been difficult to reliably amplify from the diversity of fungi either due to variability at primer binding sites or introns). ITS refers to 2 regions in the nuclear ribosomal RNA gene complex (5′ external transcribed sequence—18s rRNA—ITS1–5.8s rRNA—ITS2—28s rRNA—3′ external transcribed sequence), which is present in several thousand copies in each cell. Advantages of ITS as a barcode region include availability of broad-range primers that bind to conserved regions in 18s and 28s rRNA; presence of multiple copies per cell, facilitating recovery from small or degraded samples; and the legacy of ITS fungal sequences in GenBank. Disadvantages of ITS as barcode locus are that is a non-coding region, making it more difficult to align and compare sequences; multiple copies per cell of which may differ from one another; and presence of misidentified sequences in the legacy data.
Kelly and colleagues sampled 112 freshly collected and herbarium specimens from one genus (Usnea) including 16 of the 19 species occurring in the British Isles and 248 specimens from native woodland habitats in Britain, comprising “94 species from 55, 28, and 8 genera, families and orders, respectively.” In the latter floristic set, 66.0% of species were represented by 3 or more samples and 77.7% by 2 or more samples. DNA was extracted using DNAeasy Plant Mini Kit and amplifications were performed with sets of standard primers that amplify the entire ITS segment (ITS1-5.8s rRNA-ITS); nested PCR was performed on “a small number of samples that failed to yield a single discrete product with standard PCR.” If these failed to generate a suitable product for sequencing, then a “thin slice of a single apothecium” was placed directly into the PCR mix and amplified as above or using primers for ITS2 only. The full ITS region was obtained from 80.9% of combined 351 samples (75.9% of Usnea and 83.9% floristic). 22 (6.3%) of products showed heterogeneity on direct sequencing and required cloning to obtain suitable products for sequencing. The commonest regions for failure were no amplification [7.1% overall, largely with older (>3y) specimens; and amplification of non-target fungi (2.0% overall, only with field samples from floristic dataset)].
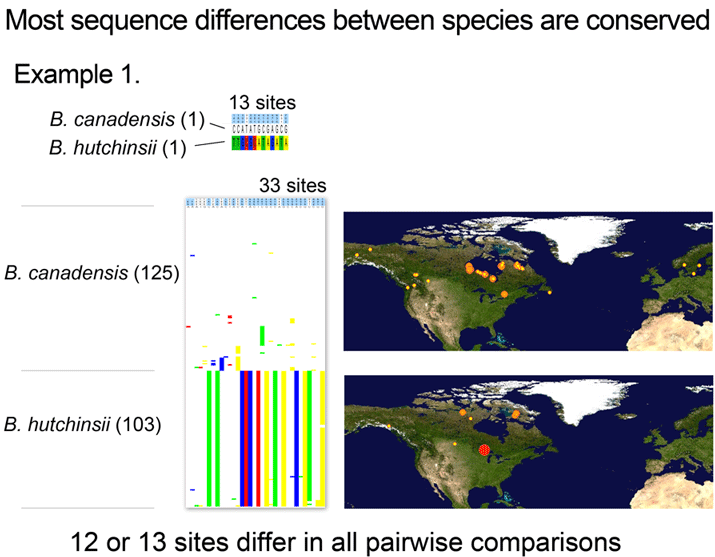
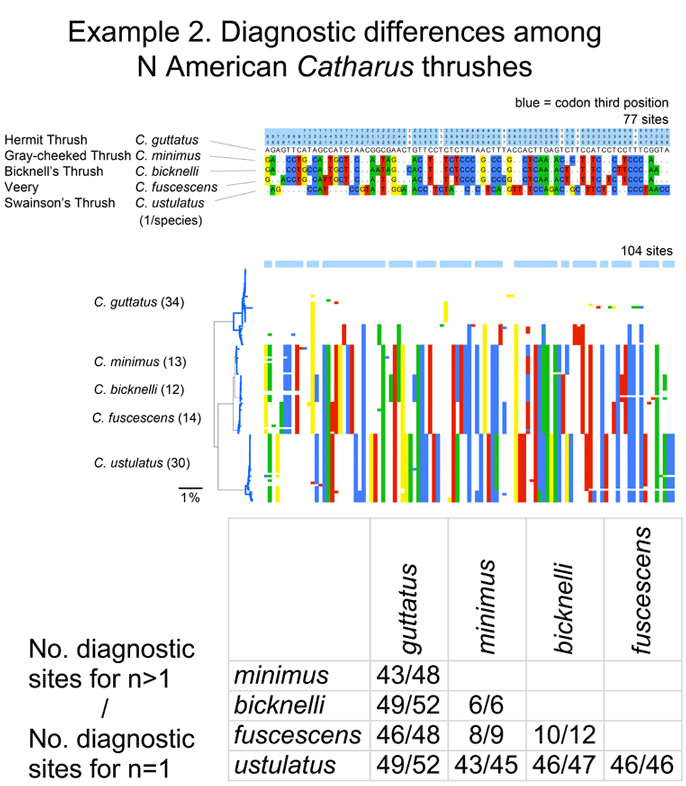
Is there a “barcode gap” (intraspecific<<interspecific distance) among fungal ITS sequences? In this study at least, usually yes. The RBG researchers defined clusters as nodes with ≥ 70 BP under BIONJ method or PP ≥ 0.95 under Bayesian inference. Under these criteria, species discrimination was 73.3% for Usnea dataset and 92.1% for floristic dataset. Simple BLAST analysis was also usually accurate–80% of Usnea species and 92.1 of floristic species were correctly assigned. This bodes well for cataloging the “dark matter” of fungal biodiversity using ITS DNA barcodes. So little is now known, it is exciting to contemplate what will be learned!