Breath tests for DNA
In August 2010 PLoS ONE, researchers from University of Queensland, Georgetown University, and National Aquarium look at feasibility of genotyping cetaceans (whales, dolphins, and porpoises) by sampling blow, the exhalations from blowholes. The standard method for collecting cetacean DNA, dart biopsying, is considered inappropriate in some settings, particularly for young animals. Blow sampling has been used to assess disease in free-ranging cetaceans (Acevedo-Whitehouse et al Anim Cons 2009).
In the PLoS ONE report, Frère and colleagues studied six bottlenose dolphins (Tursiops truncatus) housed at the National Aquarium from which they were able to collect both blood and blow samples. Blow sampling involved holding a 50 mL polypropylene tube inverted over the blowhole of “dolphins trained to exhale on cue.” Tubes were placed on dry ice for transport to the laboratory, where the presumably adherent blow material was resuspended in 500 μL of TE buffer (this worked better than ethanol), and centrifuged at 3000 rpm for 3 min. Excess TE was removed, and DNA was extracted using a Qiagen DNeasy Blood and Tissue Kit. For all six individuals, mitochondrial and microsatellite DNA profiles from blow matched those from blood. The researchers applied this approach to a wild population of bottlenose dolphins in the eastern gulf of Shark Bay, Australia, using “a modified embroidery hoop with sterile filter paper stretched over its centre,” with successful recovery of mitochondrial DNA from one individual so far. 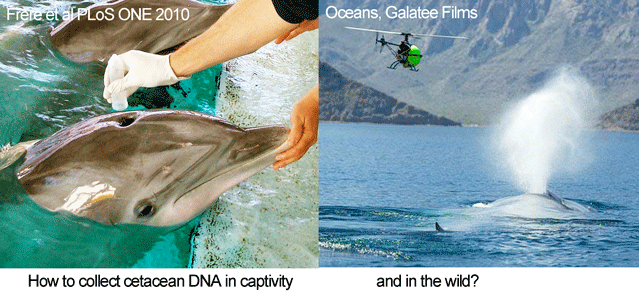
Looking ahead, small, remote-controlled devices might be used for sampling, as were employed in filming cetaceans in Oceans. There may also be applications of DNA breath-testing in  land animals (see Schlieren image of extensive turbulent flow following a cough). More generally, the increasing sensitivity of DNA techniques opens a dizzying array of possibilities for DNA-based identification. For example, forensic laboratories now routinely employ “touch DNA” methods sensitive enough to detect the tiny number of cells that are routinely shed when we touch objects, and the presence of amphibians in a pond can be determined by DNA testing a 15 mL water sample (Ficetola Biol Lett 2008).
land animals (see Schlieren image of extensive turbulent flow following a cough). More generally, the increasing sensitivity of DNA techniques opens a dizzying array of possibilities for DNA-based identification. For example, forensic laboratories now routinely employ “touch DNA” methods sensitive enough to detect the tiny number of cells that are routinely shed when we touch objects, and the presence of amphibians in a pond can be determined by DNA testing a 15 mL water sample (Ficetola Biol Lett 2008).
This entry was posted on Tuesday, January 18th, 2011 at 3:32 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.