Taxonomy needs DNA, and quick, simple ways to analyze it
 Lumpsuckers are globular, scaleless marine fish with bony tubercles on head and body, and a ventral sucking disc, derived from specialized pelvic fins, which allows them to adhere to environmental substrates. The genus Eumicrotremus comprises 16 species distributed in the Arctic and northern Atlantic and Pacific oceans; the commonest and most widespread in the north Atlantic is the Spiny lumpsucker E. spinosus, which was first described by Fabricius in 1776. A new subspecies E. s. eggvinii was described in 1956, based on a single specimen, and this was later elevated to species level “on the basis of wrinkled skin, numerous dermal warts and a large sucking disk, in addition to the low number of bony tubercles.”
Lumpsuckers are globular, scaleless marine fish with bony tubercles on head and body, and a ventral sucking disc, derived from specialized pelvic fins, which allows them to adhere to environmental substrates. The genus Eumicrotremus comprises 16 species distributed in the Arctic and northern Atlantic and Pacific oceans; the commonest and most widespread in the north Atlantic is the Spiny lumpsucker E. spinosus, which was first described by Fabricius in 1776. A new subspecies E. s. eggvinii was described in 1956, based on a single specimen, and this was later elevated to species level “on the basis of wrinkled skin, numerous dermal warts and a large sucking disk, in addition to the low number of bony tubercles.”
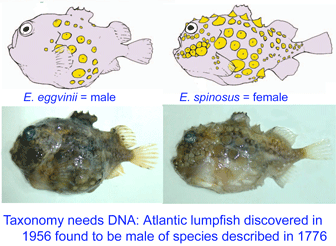 In August 2007 J Fish Biol 71A: 111, researchers from University of Bergen, Norway, analyze DNA and morphologic characters of E. eggvinii (n=16) and E. spinosus (n=67). All specimens were easily classified by morphologic characters. However, the two species had identical mitochondrial DNA sequences (COI barcode region, COII, cytb) and identical nuclear gene Tmo-4C4. Further genetic testing revealed that E. eggvinii were all males, and E. spinosus were all females. The authors conclude that the two morphologically distinct “species” represent the sexually dimorphic forms of E. spinosus.
In August 2007 J Fish Biol 71A: 111, researchers from University of Bergen, Norway, analyze DNA and morphologic characters of E. eggvinii (n=16) and E. spinosus (n=67). All specimens were easily classified by morphologic characters. However, the two species had identical mitochondrial DNA sequences (COI barcode region, COII, cytb) and identical nuclear gene Tmo-4C4. Further genetic testing revealed that E. eggvinii were all males, and E. spinosus were all females. The authors conclude that the two morphologically distinct “species” represent the sexually dimorphic forms of E. spinosus.
In this study by Byrkjedal et al, identical mtDNA sequences suggested synonymy, and this in turn suggested that morphologic divergence might represent sexual dimorphism, confirmed by further genetic testing. To my reading, this study suggests DNA testing needs to be as commonplace in taxonomy as recording size, shape, and coloration, and counting rays in fins and placement of tubercles. Every new species should have a representative DNA sequence as part of the species description. For animals, the standard should be a COI barcode. One of the remaining impediments to widespread adoption is that simple protocols for sequencing COI barcode region need to be better disseminated. In this study, the researchers were able to recover COI barcode region using primers designed for invertebrates (Folmer et al 1994), although others have published primer pairs that have greatly increased effectiveness with diverse fish (Ward et al 2005, Ivanova et al 2007). Compiling primer pairs and amplification protocols and displaying this information prominently on the various barcoding web sites will help (see for example SpongeBOL home page www.spongebarcoding.org link to illustrated primer primer!). I close with note this is post #100 since the first DNA barcode blog entry of March 15, 2006!
This entry was posted on Saturday, January 12th, 2008 at 6:29 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.