1/1000 animal diversity mapped
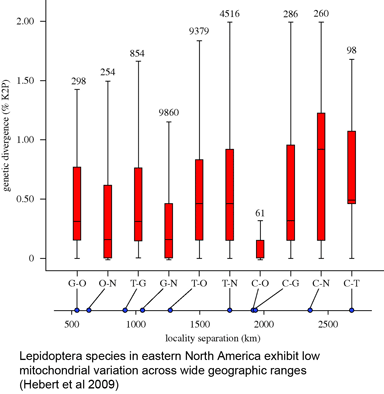 In 16 December 2009 Biol Lett, researchers from University of Guelph, University of British Columbia, and Agriculture and Agri-Food Canada report on COI barcodes for 11,289 individuals representing 1,327 species of Lepidoptera (moths and butterflies) collected in eastern North America. This large collection revealed the same patterns of highly restricted intraspecific variation, uncommon barcode sharing, and overlooked diversity seen in numerous smaller studies: average variation within species was 0.43%, while average among congeneric species was 7.7%, 18-fold higher. Only nine cases (0.7%) of barcode sharing between species were observed, and at the same time, large divergences (>2%) suggesting overlooked taxa were found in 67 (5.1%) of cases (in some cases morphological and ecological differences supporting species status were observed). The survey included multiple individuals per species collected at sites 500 to 2800 km apart, with “no significant increase in genetic distances with geographical separation.” Hebert, deWaard, and Landry conclude “an effective identification system can be constructed for the Lepidoptera fauna of eastern North American without extensive geographical surveys of each species,” and that, given likely similar patterns in most terrestrial and marine fauna, “a comprehensive barcode library for animal life can be assembled rapidly,” with diverse benefits to society and science.
In 16 December 2009 Biol Lett, researchers from University of Guelph, University of British Columbia, and Agriculture and Agri-Food Canada report on COI barcodes for 11,289 individuals representing 1,327 species of Lepidoptera (moths and butterflies) collected in eastern North America. This large collection revealed the same patterns of highly restricted intraspecific variation, uncommon barcode sharing, and overlooked diversity seen in numerous smaller studies: average variation within species was 0.43%, while average among congeneric species was 7.7%, 18-fold higher. Only nine cases (0.7%) of barcode sharing between species were observed, and at the same time, large divergences (>2%) suggesting overlooked taxa were found in 67 (5.1%) of cases (in some cases morphological and ecological differences supporting species status were observed). The survey included multiple individuals per species collected at sites 500 to 2800 km apart, with “no significant increase in genetic distances with geographical separation.” Hebert, deWaard, and Landry conclude “an effective identification system can be constructed for the Lepidoptera fauna of eastern North American without extensive geographical surveys of each species,” and that, given likely similar patterns in most terrestrial and marine fauna, “a comprehensive barcode library for animal life can be assembled rapidly,” with diverse benefits to society and science.
In my view, this study should lay to rest the early and persisting worries of some taxonomists that single gene DNA barcoding would distinguish species only in limited situations. For example, in a 2004 PLoS Biol commentary, Moritz and Cicero cautioned “But to determine when and where [DNA barcoding]…is applicable, we now need to discover the boundary conditions.” The 2009 answer is that there are no major restrictions to wide application of DNA barcoding in animals, taxonomically or geographically, and the one regularly encountered limitation is very young species, which represent a small fraction of recognized taxa even in intensively studied groups. At the same time DNA barcoding speeds taxonomic assessment by flagging genetically distinct forms, many of which are found represent unrecognized species, including species that would likely otherwise remain hidden indefinitely. Together with prior work this study refutes a widely-cited (pre-barcoding) estimate that 23% of animal taxa have shared or overlapping mitochondrial DNA sequences (Funk and Omland Ann Rev Genet 2003); this estimate presumably reflected biases in then-existing databases. In closing, I note that Hebert, deWaard, and Landry offer a new yardstick, namely the fraction of the animal kingdom mapped, lifting our eyes up to the goal of a rapid identification system for all eukaryotic life.
This entry was posted on Thursday, December 24th, 2009 at 3:49 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.