Dinoflagellate diversity revealed by DNA
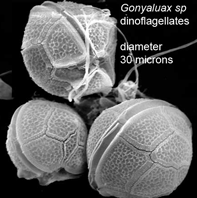 Peering into the vast diversity of life beyond multicellular eukaryotes (animals, plants, and fungi) is dizzying. In March 2009 Applied Environ Microbiol researchers from University of Connecticut assess dinoflagellate diversity with mitochondrial DNA sequencing. Dinoflagellates are unicellular, often photosynthetic, mostly marine plankton characteristically having two flagella and encased in a segmented hardened exterior. Dinoflagellate blooms are the cause of red tides, and dinoflagellate toxins ingested by fish and shellfish are the cause of ciguatera and paralytic shellfish poisoning. For unknown reasons, some species are bioluminescent when mechanically stimulated, producing glowing displays when perturbed by waves, fish, or kayakers, for example.
Peering into the vast diversity of life beyond multicellular eukaryotes (animals, plants, and fungi) is dizzying. In March 2009 Applied Environ Microbiol researchers from University of Connecticut assess dinoflagellate diversity with mitochondrial DNA sequencing. Dinoflagellates are unicellular, often photosynthetic, mostly marine plankton characteristically having two flagella and encased in a segmented hardened exterior. Dinoflagellate blooms are the cause of red tides, and dinoflagellate toxins ingested by fish and shellfish are the cause of ciguatera and paralytic shellfish poisoning. For unknown reasons, some species are bioluminescent when mechanically stimulated, producing glowing displays when perturbed by waves, fish, or kayakers, for example.
As a first step toward creating a reference library, Lin and colleagues compiled mtDNA sequences from 49 dinoflagellate species representing six orders (this included 20 COI and 60 cytochrome b sequences; 12 of the latter were newly obtained in this study). As there are about 2500 named dinoflagellate species, this is a sparsely-populated reference library so far. In addition, there were multiple samples from just 5 species, so intraspecific variation is not yet well-studied. As an aside, I note that most of the published and new sequences were derived from strains maintained at Pravasoli-Guillard National Center for Culture of Marine Phytoplankton (CCMP). There is no explicit mention of CCMP in the paper or GenBank depositions, although a plankton specialist would probably recognize the source from sample designations. More generally, there is no formal documentation of taxonomic identifications (eg collection sources for cultures or photographs for environmental samples and/or individual who performed identifications). Although this is not unusual in taxonomic papers, it seems to me that identifications should be as well documented as for example PCR conditions.
In preparing the reference library, the researchers were unable to develop primers that amplified the barcode region of COI efficiently (ie the primers worked with some species and not others) and instead focused on cytochrome b using a primer pair that amplified a 385 bp segment. The primer difficulty is surprising given that COI is usually more conserved than cyt b (including in dinoflagellates), which should make it easier to design broad-range primers. 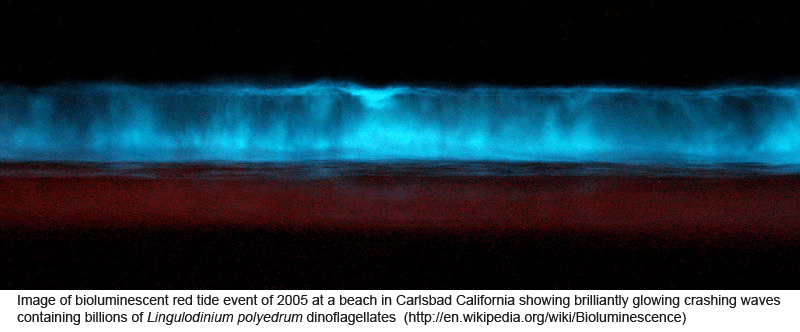
The researchers then analyzed pooled environmental DNA samples prepared by filtering water specimens collected during different months at 3 marine stations in Long Island Sound and at a freshwater retention pond (Mirror Lake) on the University of Connecticut campus. While PCR products from monospecific cultures were sequenced directly, those from environmental samples were first cloned, and then 20 to 50 clones from each water sample were sequenced (total clones analyzed 450).
Lin and co-workers obtained a large number of distinct haplotypes from the environmental samples; by my inspection of their phylogram nearly all of the clones (>420) were unique. Only a small minority could be assigned to known species or genera. On the technical side, the authors used a complex model of nucleotide substitution (TVM+G) to calculate differences among haplotypes and UPGMA to create trees, so their distance results and trees are not directly comparable to those in most DNA barcoding papers, which use K2P- or p-distances to calculate differences and neighbor-joining to create trees. In any case, according to the authors, the sequence results consistently showed greater diversity than was detected through microscopic analysis, “likely caused by the much higher detection sensitivity of PCR than of microscopic counting and by some genotypes that could not be discriminated morphologically.” The authors conclude “[w]hen a broader cob [cyt b] database becomes available, the taxon-resolving power of this gene would certainly increase.” I hope they or others will also develop efficient primer sets for amplifying COI in addition to cyt b.
Looking ahead, the reference library can be augmented relatively inexpensively by analyzing mtDNA sequences of the 2400 strains at CCMP. However, the mtDNA diversity in this study suggests dozens of new species from just 4 sampling sites around Connecticut, implying the global total of undescribed species is very large. This suggests a need for some sort of “automated species identifier”: a machine approach that would sort samples into individual cells, then photograph, sequence, apply MOTU-type analysis, for example. In the meantime, it may be necessary to work with pooled sequences from environmental samples, as is done for bacterial communities, without attempting to delineate species.
This entry was posted on Sunday, April 12th, 2009 at 7:06 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.
April 14th, 2009 at 4:55 am
More generally, there is no formal documentation of taxonomic identifications (eg collection sources for cultures or photographs for environmental samples and/or individual who performed identifications). Although this is not unusual in taxonomic papers, it seems to me that identifications should be as well documented as for example PCR conditions.
I am not familiar with papers in microorganisms taxonomy, but in entomology (arguably also a very diverse and species-rich group of organisms) if you don’t document identification extensively you don’t get pass the peer-review process.
So yes, the community (e.g., reviewers) of DNA barcoding research should enforce this as soon as possible.