Freshwater fish DNA data debut
Sunday, June 22nd, 2008
 In June 2008 PLoS ONE, thirteen researchers from nine Canadian universities, museums, and federal agencies report on mtDNA sequences from 1360 individuals representing 195 (95%) of Canada’s 205 freshwater fish species. Hubert et al follow “best practices” established for DNA barcode records (similar criteria would enhance the value of other genetic reference data as well), namely each sequence is derived from a vouchered specimen and the barcode record includes:
In June 2008 PLoS ONE, thirteen researchers from nine Canadian universities, museums, and federal agencies report on mtDNA sequences from 1360 individuals representing 195 (95%) of Canada’s 205 freshwater fish species. Hubert et al follow “best practices” established for DNA barcode records (similar criteria would enhance the value of other genetic reference data as well), namely each sequence is derived from a vouchered specimen and the barcode record includes:
- “Bi-directional sequences of at least 500 base-pairs from the approved barcode region of COI, containing no ambiguous sites
- Links to electropherogram trace files available in the NCBI Trace Archive
- Sequences for the forward and reverse PCR amplification
- Species names that refer to documented names in a taxonomic publication or other documentation of the species concept used
- Links to voucher specimens using the approved format of institutional acronym:collection code:catalog ID number”
The researchers analyzed an average of 7.6 specimens/species, with an effort to sample across species ranges. A first pass look at genetic distances among and within Canadian freshwater fish shows results similar to those of other animal groups: average variation within species, 0.3%; average minimum distance between congeneric species (nearest neighbor), 8.3%; species with overlapping mtDNA sequences, 7% (4 species pairs and 1 flock of 5 species; one of the overlapping species pairs represents probable introgression. ) Five species showed divergent clusters differing by 1-2% in different parts of their geographic ranges, and 2 species showed larger divergences (3%, 7%); some or all of these might represent distinct species.
A challenge for science publishing is disseminating the large data sets that are increasingly generated. Restricting publication to only those studies with novel findings can lead to a kind of distortion, sometimes with serious consequences. The bias against negative studies, for example, is one factor contributing to the misculation of risks of medicines. As biodiversity genetics moves forward, we need ways to ensure high-quality work, receive appropriate academic credit, and disseminate results in a timely manner. PLoS ONE describes itself as “an international, peer-reviewed, open-access, online publication…that welcomes reports on primary research from any scientific discipline.” It seems to me that this sort of forum with a focus on quality rather than novelty is needed as a home for publication of large genetic data sets including DNA barcode records. Making this information available in a timely manner will in turn help drive development of analytic and display tools and enable scientific applications, such as identification of fish eggs and larva shown above.
 How many giraffes were onboard the Ark? Giraffes are classified as a single species, Giraffa camelopardalis, with five to nine subspecies proposed based on regional variation in pelage (coat pattern). In
How many giraffes were onboard the Ark? Giraffes are classified as a single species, Giraffa camelopardalis, with five to nine subspecies proposed based on regional variation in pelage (coat pattern). In 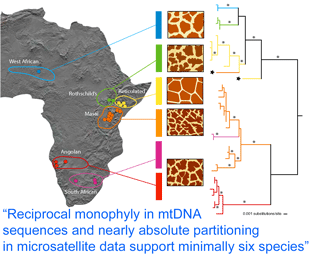 Analysis of 14 nuclear microsatellites from 381 individuals at 18 locations (it is not clear whether these are the same individuals as above) recovered the same six groups and suggested additional genetic subdivisions within some groups. Although at least some of the genetically and pelage-defined clusters have overlapping or adjacent ranges without geographic barriers, only three (0.8%) of individuals were identified as hybrids. These findings raise interesting questions about giraffe biology; for example, is there behavioral isolation perhaps based on visual recognition of pelage patterns?
Analysis of 14 nuclear microsatellites from 381 individuals at 18 locations (it is not clear whether these are the same individuals as above) recovered the same six groups and suggested additional genetic subdivisions within some groups. Although at least some of the genetically and pelage-defined clusters have overlapping or adjacent ranges without geographic barriers, only three (0.8%) of individuals were identified as hybrids. These findings raise interesting questions about giraffe biology; for example, is there behavioral isolation perhaps based on visual recognition of pelage patterns? 
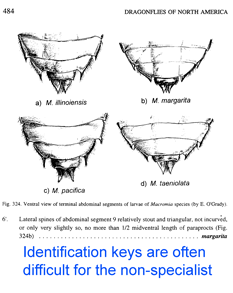 Just two years ago,
Just two years ago,