Revealing krill diets with DNA
 Krill are shrimp-like crustaceans found throughout the world’s oceans. The Antarctic krill, Euphasia superba, is thought to be the most abundant species on the planet in terms of biomass (500 million metric tonnes corresponding to 5 x 10^14 individuals), is a primary food source for whales, seals, and oceanic birds, and functions as a major planetary carbon sink by excreting waste that sinks to ocean floor. What does this very abundant, central-to-food-web species eat? For many animals, observation of eating behavior is impractical, and analysis of stomach contents or feces may be the only way to determine diet. However, such material may be morphologically unrecognizable.
Krill are shrimp-like crustaceans found throughout the world’s oceans. The Antarctic krill, Euphasia superba, is thought to be the most abundant species on the planet in terms of biomass (500 million metric tonnes corresponding to 5 x 10^14 individuals), is a primary food source for whales, seals, and oceanic birds, and functions as a major planetary carbon sink by excreting waste that sinks to ocean floor. What does this very abundant, central-to-food-web species eat? For many animals, observation of eating behavior is impractical, and analysis of stomach contents or feces may be the only way to determine diet. However, such material may be morphologically unrecognizable.
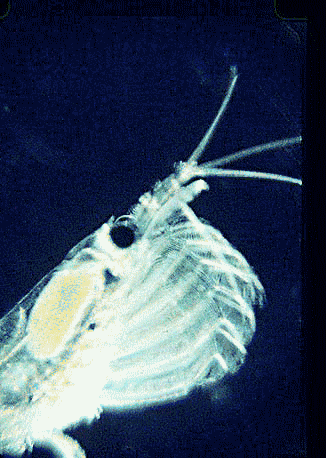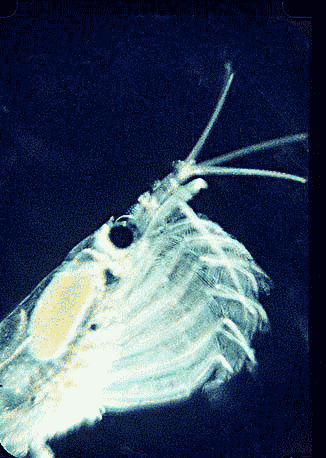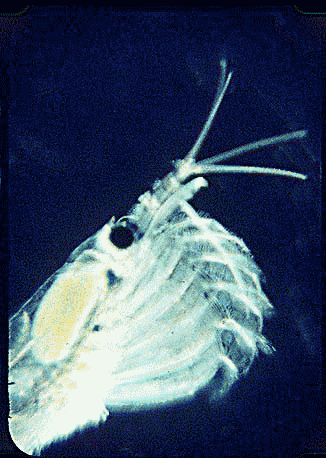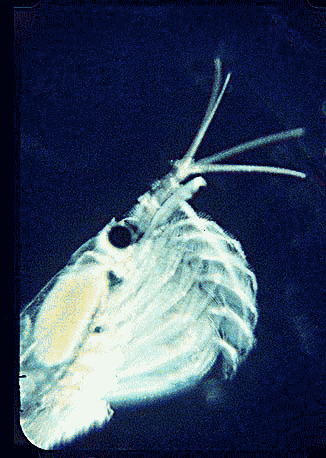 In August 2006 Marine Biotech 8:686, researchers from University of Tasmania and Department of Environment, Tasmania, compare DNA sequencing and light microscopy in identifying prey in stomach contents of E. superba. Passmore et al isolated DNA from stomach contents of 6 ethanol preserved krill and, using diatom-specific primers, amplified a 103 bp portion of nuclear small subunit RNA (ssRNA). ssRNA was used because at present it has the best taxonomic representation in GenBank for krill prey species. The researchers sequenced at least 50 clones from each individual krill and found 14 OTUs (operational taxonomic units), with 86% to 100% match to GenBank sequences. These results were compared to microscopic identification of diatom silica skeleton fragments in stomach contents, which involved counting 1000-3000 fragments per individual. Results were similar, although DNA analysis and light microscopy each appeared more sensitive for certain species. This study might be a best case for light microscopy because silica-skeletoned diatoms are not easily digested. As the authors point out, krill also consume a range of protozoa and small zooplankton, and the importance of these sources may be underappreciated.The authors conclude “the application of DNA diet analysis to krill warrants further investigation, particularly for prey that are difficult to study using other methods“.
In August 2006 Marine Biotech 8:686, researchers from University of Tasmania and Department of Environment, Tasmania, compare DNA sequencing and light microscopy in identifying prey in stomach contents of E. superba. Passmore et al isolated DNA from stomach contents of 6 ethanol preserved krill and, using diatom-specific primers, amplified a 103 bp portion of nuclear small subunit RNA (ssRNA). ssRNA was used because at present it has the best taxonomic representation in GenBank for krill prey species. The researchers sequenced at least 50 clones from each individual krill and found 14 OTUs (operational taxonomic units), with 86% to 100% match to GenBank sequences. These results were compared to microscopic identification of diatom silica skeleton fragments in stomach contents, which involved counting 1000-3000 fragments per individual. Results were similar, although DNA analysis and light microscopy each appeared more sensitive for certain species. This study might be a best case for light microscopy because silica-skeletoned diatoms are not easily digested. As the authors point out, krill also consume a range of protozoa and small zooplankton, and the importance of these sources may be underappreciated.The authors conclude “the application of DNA diet analysis to krill warrants further investigation, particularly for prey that are difficult to study using other methods“.
This work shows the essential need for a comprehensive reference library, so far lacking. A study underway is examining mitochondrial and nuclear genes as barcodes for phytoplankton. Looking ahead, a “massively parallel” pyrosequencing approach could enable rapid and representative analysis of mixed environmental samples, such as stomach contents, without biases resulting from amplification and cloning.
This entry was posted on Saturday, January 27th, 2007 at 11:38 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.
December 16th, 2015 at 11:32 pm
Interesting study. Perhaps modern methods could be used to more quickly and cheaply identify the matching DNA between the stomach contents of krill and the range of protozoa and small zooplankton. It would be interesting to track how environmental changes might affect these species.